Chapter 10 TME Modeling
Previous studies have shown that the tumour microenvironment is a complex ecosystem. No single cell or gene is sufficient to influence the phenotype. Therefore, machine learning models of the tumour microenvironment or models of tumour microenvironment typing are used to predict tumour phenotypes and treatment response. In the last section, we present common considerations and scenarios for constructing tumour microenvironment models.
10.2 Data prepare
Using data from IMvigor210, we demonstrate two common scenarios for building models of the tumour microenvironment: predicting survival and predicting treatment response (BOR, RECIEST 1.1).
10.3 Input data (overall survival) prepare
pdata_prog <- imvigor210_pdata %>%
dplyr::select(ID, OS_days, OS_status) %>%
mutate(OS_days = as.numeric(.$OS_days)) %>%
mutate(OS_status = as.numeric(.$OS_status))
head(pdata_prog)## # A tibble: 6 × 3
## ID OS_days OS_status
## <chr> <dbl> <dbl>
## 1 SAM00b9e5c52da9 57.2 1
## 2 SAM0257bbbbd388 469. 1
## 3 SAM025b45c27e05 263. 1
## 4 SAM032c642382a7 74.9 1
## 5 SAM04c589eb3fb3 20.7 0
## 6 SAM0571f17f4045 136. 110.4 Constructing survival prediction models
prognostic_result <- PrognosticModel(x = imvigor210_sig,
y = pdata_prog,
scale = T,
seed = 123456,
train_ratio = 0.7,
nfold = 8,
plot = TRUE)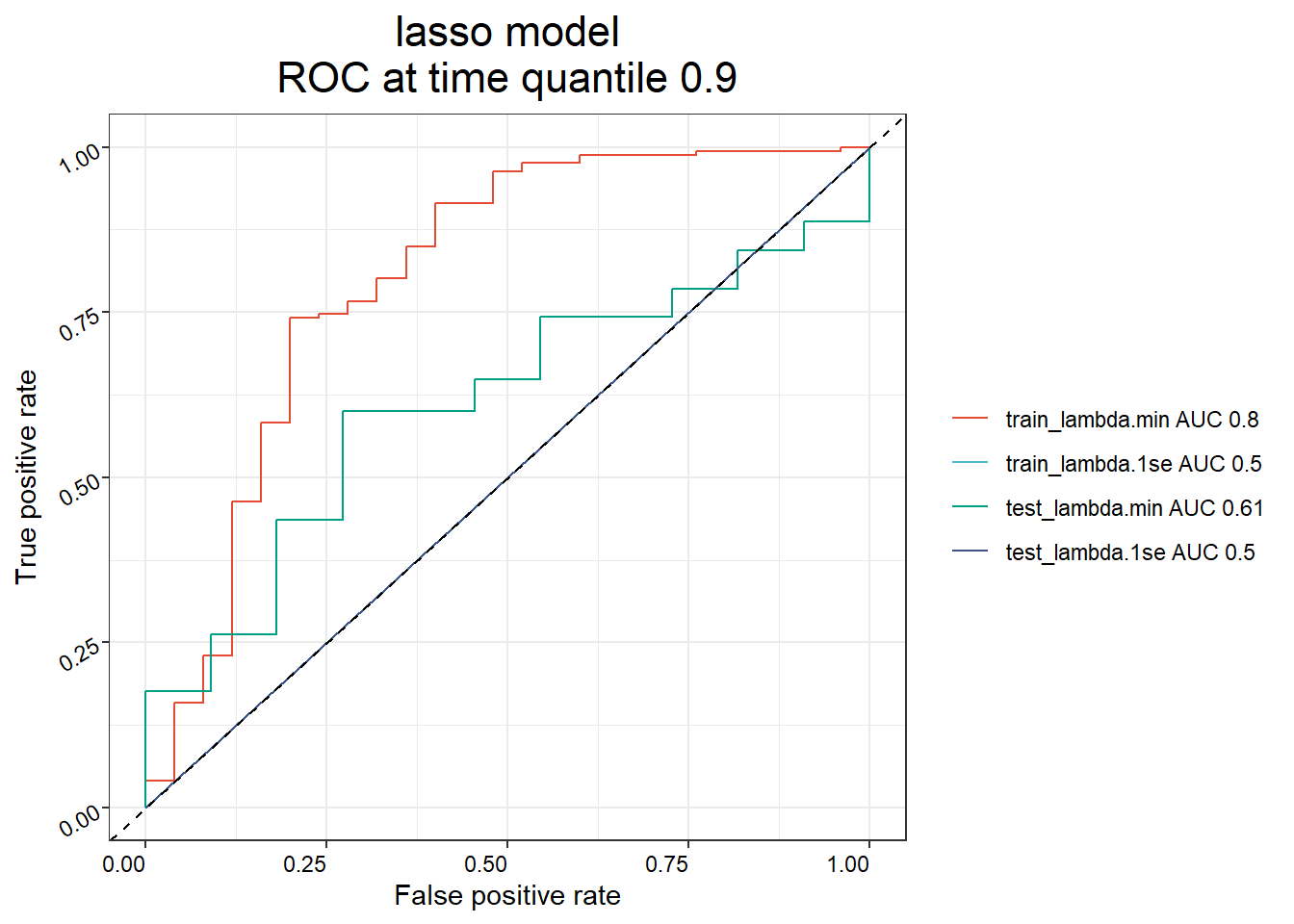
10.5 Input data (Response) prepare
pdata_group <- imvigor210_pdata[!imvigor210_pdata$BOR_binary=="NA",c("ID","BOR_binary")]
pdata_group$BOR_binary <- ifelse(pdata_group$BOR_binary == "R", 1, 0)
head(pdata_group)## # A tibble: 6 × 2
## ID BOR_binary
## <chr> <dbl>
## 1 SAM0257bbbbd388 0
## 2 SAM025b45c27e05 0
## 3 SAM032c642382a7 0
## 4 SAM0571f17f4045 0
## 5 SAM065890737112 1
## 6 SAM0684af734db1 110.6 Constructing prediction models for response
binom_res <- BinomialModel(x = imvigor210_sig,
y = pdata_group,
seed = 123456,
scale = TRUE,
train_ratio = 0.7,
nfold = 8,
plot = T)## NULL## NULL## NULL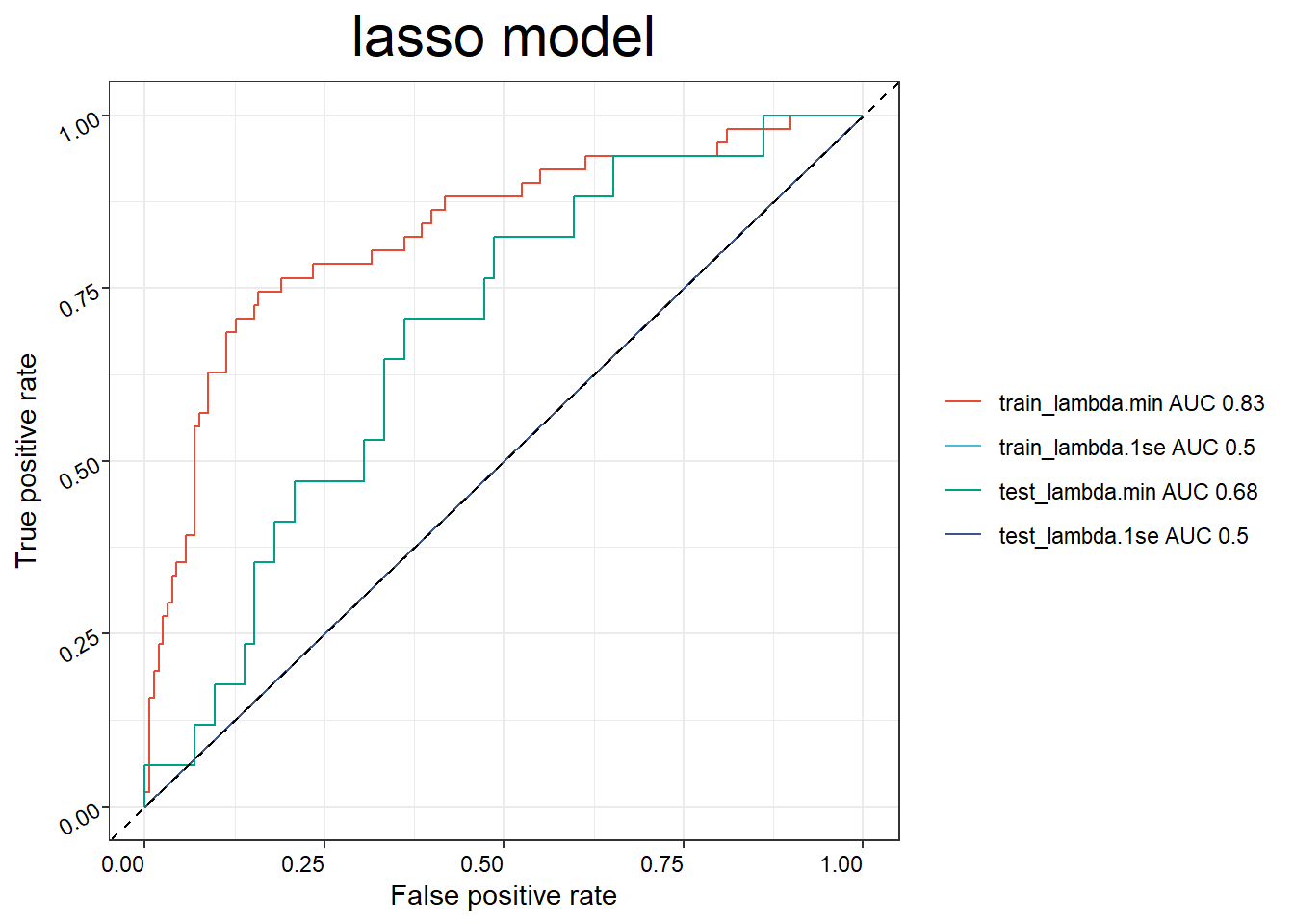
## NULL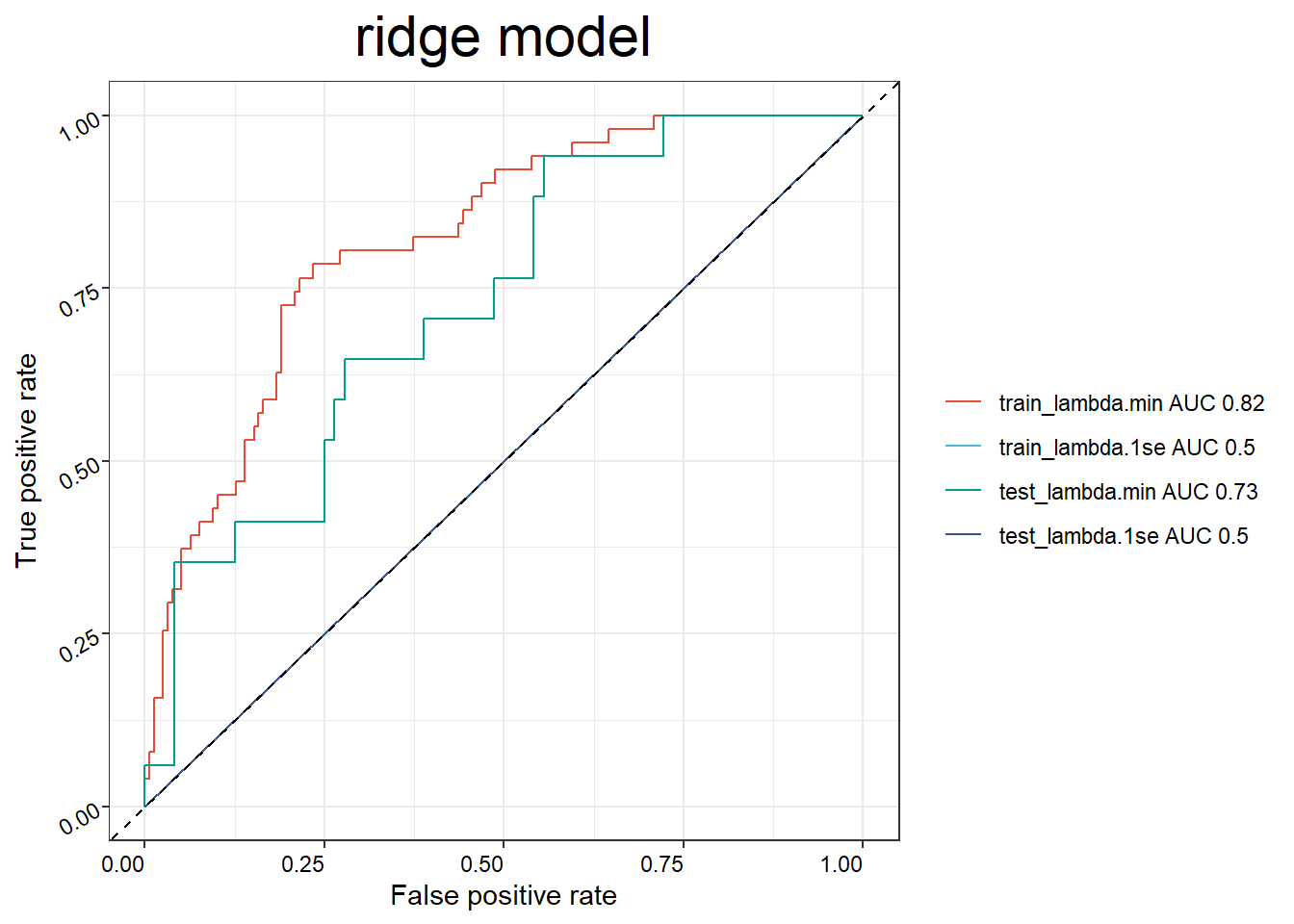
## NULL10.7 References
Cristescu, R., Lee, J., Nebozhyn, M. et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 21, 449–456 (2015). https://doi.org/10.1038/nm.3850
CIBERSORT; Newman, A. M., Liu, C. L., Green, M. R., Gentles, A. J., Feng, W., Xu, Y., … Alizadeh, A. A. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nature Methods, 12(5), 453–457. https://doi.org/10.1038/nmeth.3337;
Seurat: Hao and Hao et al. Integrated analysis of multimodal single-cell data. Cell (2021)