Chapter 8 Tumor ecosystem analysis
8.2 Downloading data for example
Obtaining data set from GEO Gastric cancer: GSE62254 using GEOquery R package.
if (!requireNamespace("GEOquery", quietly = TRUE)) BiocManager::install("GEOquery")
library("GEOquery")
# NOTE: This process may take a few minutes which depends on the internet connection speed. Please wait for its completion.
eset_geo<-getGEO(GEO = "GSE62254", getGPL = F, destdir = "./")
eset <-eset_geo[[1]]
eset <-exprs(eset)
eset[1:5,1:5]## GSM1523727 GSM1523728 GSM1523729 GSM1523744 GSM1523745
## 1007_s_at 3.2176645 3.0624323 3.0279131 2.921683 2.8456013
## 1053_at 2.4050109 2.4394879 2.2442708 2.345916 2.4328582
## 117_at 1.4933412 1.8067380 1.5959665 1.839822 1.8326058
## 121_at 2.1965561 2.2812181 2.1865556 2.258599 2.1874363
## 1255_g_at 0.8698382 0.9502466 0.8125414 1.012860 0.94419938.3 Gene Annotation: HGU133PLUS-2 (Affaymetrix)
# Conduct gene annotation using `anno_hug133plus2` file; If identical gene symbols exists, these genes would be ordered by the mean expression levels. The gene symbol with highest mean expression level is selected and remove others.
eset<-anno_eset(eset = eset,
annotation = anno_hug133plus2,
symbol = "symbol",
probe = "probe_id",
method = "mean")
eset[1:5, 1:3]## GSM1523727 GSM1523728 GSM1523729
## SH3KBP1 4.327974 4.316195 4.351425
## RPL41 4.246149 4.246808 4.257940
## EEF1A1 4.293762 4.291038 4.262199
## COX2 4.250288 4.283714 4.270508
## LOC101928826 4.219303 4.219670 4.2132528.4 Determine TME subtype of gastric cancer using TMEclassifier R package
if (!requireNamespace("TMEclassifier", quietly = TRUE)) devtools::install_github("LiaoWJLab/TMEclassifier")
library(TMEclassifier)
tme <- tme_classifier(eset = eset, scale = TRUE)## Step-1: Expression data preprocessing...## Step-2: TME deconvolution...## Step-3: Predicting TME phenotypes...## [09:52:29] WARNING: src/learner.cc:1203:
## If you are loading a serialized model (like pickle in Python, RDS in R) generated by
## older XGBoost, please export the model by calling `Booster.save_model` from that version
## first, then load it back in current version. See:
##
## https://xgboost.readthedocs.io/en/latest/tutorials/saving_model.html
##
## for more details about differences between saving model and serializing.
##
## [09:52:29] WARNING: src/learner.cc:888: Found JSON model saved before XGBoost 1.6, please save the model using current version again. The support for old JSON model will be discontinued in XGBoost 2.3.
## [09:52:29] WARNING: src/learner.cc:553:
## If you are loading a serialized model (like pickle in Python, RDS in R) generated by
## older XGBoost, please export the model by calling `Booster.save_model` from that version
## first, then load it back in current version. See:
##
## https://xgboost.readthedocs.io/en/latest/tutorials/saving_model.html
##
## for more details about differences between saving model and serializing.## >>>--- DONE!##
## IA IE IS
## 107 96 97## ID IE IS IA TMEcluster
## 1 GSM1523727 0.204623557 0.11212681 0.68324962 IA
## 2 GSM1523728 0.009599504 0.11179146 0.87860903 IA
## 3 GSM1523729 0.852615046 0.11369089 0.03369407 IE
## 4 GSM1523744 0.053842233 0.06994632 0.87621145 IA
## 5 GSM1523745 0.055973019 0.80839488 0.13563209 IS
## 6 GSM1523746 0.545343299 0.37437568 0.08028102 IE##
## IA IE IS
## 107 96 97## ID IE IS IA TMEcluster
## 1 GSM1523727 0.204623557 0.11212681 0.68324962 IA
## 2 GSM1523728 0.009599504 0.11179146 0.87860903 IA
## 3 GSM1523729 0.852615046 0.11369089 0.03369407 IE
## 4 GSM1523744 0.053842233 0.06994632 0.87621145 IA
## 5 GSM1523745 0.055973019 0.80839488 0.13563209 IS
## 6 GSM1523746 0.545343299 0.37437568 0.08028102 IE8.5 DEG analysis: method1
Differential analysis of selected immune-activated and immune-expelled gastric cancers
pdata <- tme[!tme$TMEcluster=="IS", ]
deg <- iobr_deg(eset = eset,
annotation = NULL,
pdata = pdata,
group_id = "TMEcluster",
pdata_id = "ID",
array = TRUE,
method = "limma",
contrast = c("IA","IE"),
path = "result",
padj_cutoff = 0.01,
logfc_cutoff = 0.5)## >>>== Matching grouping information and expression matrix## >>>== limma was selected for differential gene analysis of Array data## group1 = IE## group2 = NA## # A tibble: 6 × 11
## symbol log2FoldChange AveExpr t pvalue padj B sigORnot label
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 TMEM100 0.774 1.84 13.9 2.47e-31 5.37e-27 60.4 Up_regulat… Both
## 2 ABCA8 0.933 1.90 12.9 3.11e-28 3.38e-24 53.4 Up_regulat… Both
## 3 HHIP 0.613 1.73 12.1 7.62e-26 4.46e-22 48.0 Up_regulat… Both
## 4 LMNB2 -0.287 2.25 -12.1 9.28e-26 4.46e-22 47.8 NOT Sign…
## 5 MCM6 -0.211 3.02 -12.1 1.02e-25 4.46e-22 47.7 NOT Sign…
## 6 ADH1B 0.907 1.86 12.0 2.27e-25 7.04e-22 47.0 Up_regulat… Both
## # ℹ 2 more variables: IE <dbl>, `` <dbl>8.6 GSEA analysis based on differential express gene analysis results
Select the gene set list in IOBR’s signature collection.
## # A tibble: 6 × 11
## symbol log2FoldChange AveExpr t pvalue padj B sigORnot label
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <chr>
## 1 TMEM100 0.774 1.84 13.9 2.47e-31 5.37e-27 60.4 Up_regulat… Both
## 2 ABCA8 0.933 1.90 12.9 3.11e-28 3.38e-24 53.4 Up_regulat… Both
## 3 HHIP 0.613 1.73 12.1 7.62e-26 4.46e-22 48.0 Up_regulat… Both
## 4 LMNB2 -0.287 2.25 -12.1 9.28e-26 4.46e-22 47.8 NOT Sign…
## 5 MCM6 -0.211 3.02 -12.1 1.02e-25 4.46e-22 47.7 NOT Sign…
## 6 ADH1B 0.907 1.86 12.0 2.27e-25 7.04e-22 47.0 Up_regulat… Both
## # ℹ 2 more variables: IE <dbl>, `` <dbl>sig_list <- signature_collection[c("TMEscoreB_CIR", "TMEscoreA_CIR", "DNA_replication", "Base_excision_repair",
"Pan_F_TBRs", "TGFb.myCAF", "Ferroptosis", "TLS_Nature", "Glycolysis")]
sig_list## $TMEscoreB_CIR
## [1] "DCN" "SEPP1" "ACTA2" "SPARCL1" "BEX3"
## [6] "MYLK" "AKR1C1" "TIMP2" "MXRA7" "C11orf96"
## [11] "CAV1" "PDGFRA" "FHL1" "MGP" "EID1"
## [16] "LOC101930400" "DST" "GREM1" "FERMT2" "TNC"
## [21] "CYBRD1" "LTBP1" "ACTG2" "TMEM47" "SERPINE2"
## [26] "ANTXR2" "GNG11" "TAGLN" "GSTA4" "PKIG"
## [31] "MAOA" "PTRF" "FAM3B" "PBX1" "WLS"
## [36] "SELM" "SVIL" "MYH11" "AGT" "SPON1"
## [41] "TGFB1I1" "PDLIM3" "PDK4" "SYNPO2" "MSRB3"
## [46] "PROS1" "EDNRA" "AKAP12" "PSD3" "TNS1"
## [51] "JAM3" "PDZRN3" "DDR2" "HMGCS2" "SGCE"
## [56] "MRVI1" "WFDC1" "FBLN1" "FMO5" "MAOB"
## [61] "AMOTL1" "AKT3" "CNRIP1" "CPE" "MAP1B"
## [66] "RBP1" "GNAI1" "FOXF2" "SORBS2" "ZCCHC24"
## [71] "ZNF704" "ARMCX1" "DIXDC1" "SSTR1" "THRB"
## [76] "C3orf70" "PKIB" "CNN1" "SYTL5" "DACT1"
## [81] "SYNPO" "GAS1" "DPYSL3" "CCDC80" "TSPYL5"
## [86] "DCHS1" "SOBP" "AOC3" "NDN" "FGF7P3"
## [91] "SMAD9" "MCC" "CLMP" "MYL9" "RBP4"
## [96] "PLN" "SPOCK1" "COL14A1" "CRYAB" "SRPX"
## [101] "EML1" "RERG" "PPP1R3C" "LOC100506718" "CH25H"
## [106] "HSPB8" "PID1" "TTC28" "STON1" "ABCG2"
## [111] "ZSCAN18" "SCIN" "C14orf132" "TMEM55A" "WASF3"
## [116] "PAPLN" "COLEC12" "ACKR1" "TMEM150C" "RAI2"
## [121] "TSPAN7" "MRGPRF" "ABCA8" "CHIC1" "NBEA"
## [126] "FAM13C" "SETBP1" "LDOC1" "TMEM100" "LOC101930349"
## [131] "PRICKLE2" "TSPAN18" "FABP4" "ARHGEF26" "ERICH5"
## [136] "MYOCD" "BEX2" "PPP1R14A" "FGF13" "RUNX1T1"
## [141] "MAGI2-AS3" "LINC01279" "REEP1" "PLAC9" "MYEF2"
## [146] "PRKD1" "RGN" "CLDN11" "ANK2" "ESRRG"
## [151] "SYNC" "ZNF667-AS1" "FGF7" "SFRP1" "HMCN1"
## [156] "TCEAL7" "OGN" "MAGI2" "MIR100HG" "FILIP1"
## [161] "LOC100507334" "ANKRD6" "PLEKHH2" "ZNF542P" "ARMCX4"
## [166] "NOV" "DCLK1" "ARHGAP28" "C2orf40" "TRHDE"
## [171] "EPHA7" "SCRG1" "ZNF677" "ZFPM2" "PEG3"
## [176] "SERP2" "ZNF415" "MAMDC2" "RBM24" "MEOX2"
##
## $TMEscoreA_CIR
## [1] "HLA-DPB1" "UBD" "LOC100509457" "WARS"
## [5] "TAP1" "HLA-DMA" "TRIM22" "PSAT1"
## [9] "CXCL10" "SOCS3" "CXCL9" "PBK"
## [13] "CCL4" "CCL5" "BCL2A1" "TRBC1"
## [17] "IDO1" "NFE2L3" "CCL3L3" "DTL"
## [21] "MMP9" "SLC2A3" "ZNF367" "RCC1"
## [25] "STIL" "TRAC" "HELLS" "GZMB"
## [29] "RTEL1-TNFRSF6B" "CXCL11" "GBP5" "CD2"
## [33] "CDCA2" "CDT1" "TNFAIP2" "TYMP"
## [37] "MICB" "SLC2A14" "GZMK" "CD8A"
## [41] "CENPH" "MND1" "BATF2" "BRIP1"
## [45] "E2F7" "KIF18A" "AIM2" "ETV7"
## [49] "ITK" "GNLY" "GPR171" "WDHD1"
## [53] "GBP4" "MB21D1" "NLRP3" "MCEMP1"
## [57] "POLR3G" "NLRC3" "KLRC2" "CLEC5A"
## [61] "ARHGAP11A" "GPR84" "IFNG" "ZBED2"
##
## $DNA_replication
## [1] "RNASEH2A" "POLD3" "DNA2" "FEN1" "POLA2" "RNASEH1"
## [7] "RPA4" "LIG1" "MCM2" "MCM3" "MCM4" "MCM5"
## [13] "MCM6" "MCM7" "PCNA" "POLE3" "POLA1" "POLD1"
## [19] "POLD2" "POLE" "POLE2" "PRIM1" "PRIM2" "POLE4"
## [25] "POLD4" "RFC1" "RFC2" "RFC3" "RFC4" "RFC5"
## [31] "RPA1" "RPA2" "RPA3" "SSBP1" "RNASEH2B" "RNASEH2C"
##
## $Base_excision_repair
## [1] "PARP2" "PARP3" "POLD3" "PARP1" "PARP4" "FEN1" "SMUG1" "NEIL2" "APEX2"
## [10] "POLL" "HMGB1" "APEX1" "LIG1" "LIG3" "MPG" "MUTYH" "NTHL1" "OGG1"
## [19] "PCNA" "POLE3" "POLB" "POLD1" "POLD2" "POLE" "POLE2" "NEIL3" "POLE4"
## [28] "POLD4" "UNG" "XRCC1" "NEIL1" "MBD4"
##
## $Pan_F_TBRs
## [1] "ACTA2" "ACTG2" "ADAM12" "ADAM19" "CNN1" "COL4A1"
## [7] "CTGF" "CTPS1" "FAM101B" "FSTL3" "HSPB1" "IGFBP3"
## [13] "PXDC1" "SEMA7A" "SH3PXD2A" "TAGLN" "TGFBI" "TNS1"
## [19] "TPM1"
##
## $<NA>
## NULL
##
## $Ferroptosis
## [1] "ACSL4" "AKR1C1-3" "ALOXs" "ATP5G3" "CARS"
## [6] "CBS" "CD44v" "CHAC1" "CISD1" "CS"
## [11] "DPP4" "FANCD2" "GCLC/GCLM" "GLS2" "GPX4"
## [16] "GSS" "HMGCR" "HSPB1/5" "KOD" "LPCAT3"
## [21] "MT1G" "NCOA4" "NFE2L2" "PTGS2" "RPL8"
## [26] "SAT1" "SLC7A11" "SQS" "TFRC" "TP53"
## [31] "TTC35/EMC2" "MESH1"
##
## $TLS_Nature
## [1] "CD79B" "CD1D" "CCR6" "LAT" "SKAP1" "CETP" "EIF1AY" "RBP5"
## [9] "PTGDS"
##
## $Glycolysis
## [1] "ACSS1" "ACSS2" "ADH1A" "ADH1B" "ADH1C" "ADH4" "ADH5"
## [8] "ADH6" "ADH7" "ADPGK" "AKR1A1" "ALDH1A3" "ALDH1B1" "ALDH2"
## [15] "ALDH3A1" "ALDH3A2" "ALDH3B1" "ALDH3B2" "ALDH7A1" "ALDH9A1" "ALDOA"
## [22] "ALDOB" "ALDOC" "BPGM" "DLAT" "DLD" "ENO1" "ENO2"
## [29] "ENO3" "FBP1" "FBP2" "G6PC" "G6PC2" "GALM" "GAPDH"
## [36] "GAPDHS" "GCK" "GPI" "HK1" "HK2" "HK3" "HKDC1"
## [43] "LDHA" "LDHAL6A" "LDHAL6B" "LDHB" "LDHC" "PANK1" "PCK1"
## [50] "PCK2" "PDHA1" "PDHA2" "PDHB" "PFKFB1" "PFKFB2" "PFKFB3"
## [57] "PFKFB4" "PFKL" "PFKM" "PFKP" "PGAM1" "PGAM2" "PGAM4"
## [64] "PGK1" "PGK2" "PGM1" "PGM2" "PKLR" "PKM" "SLC2A2"
## [71] "TPI1"gsea<- sig_gsea(deg,
genesets = sig_list,
path = "result",
gene_symbol = "symbol",
logfc = "log2FoldChange",
org = "hsa",
show_plot = FALSE,
msigdb = TRUE,
category = "H",
subcategory = NULL,
palette_bar = "set2")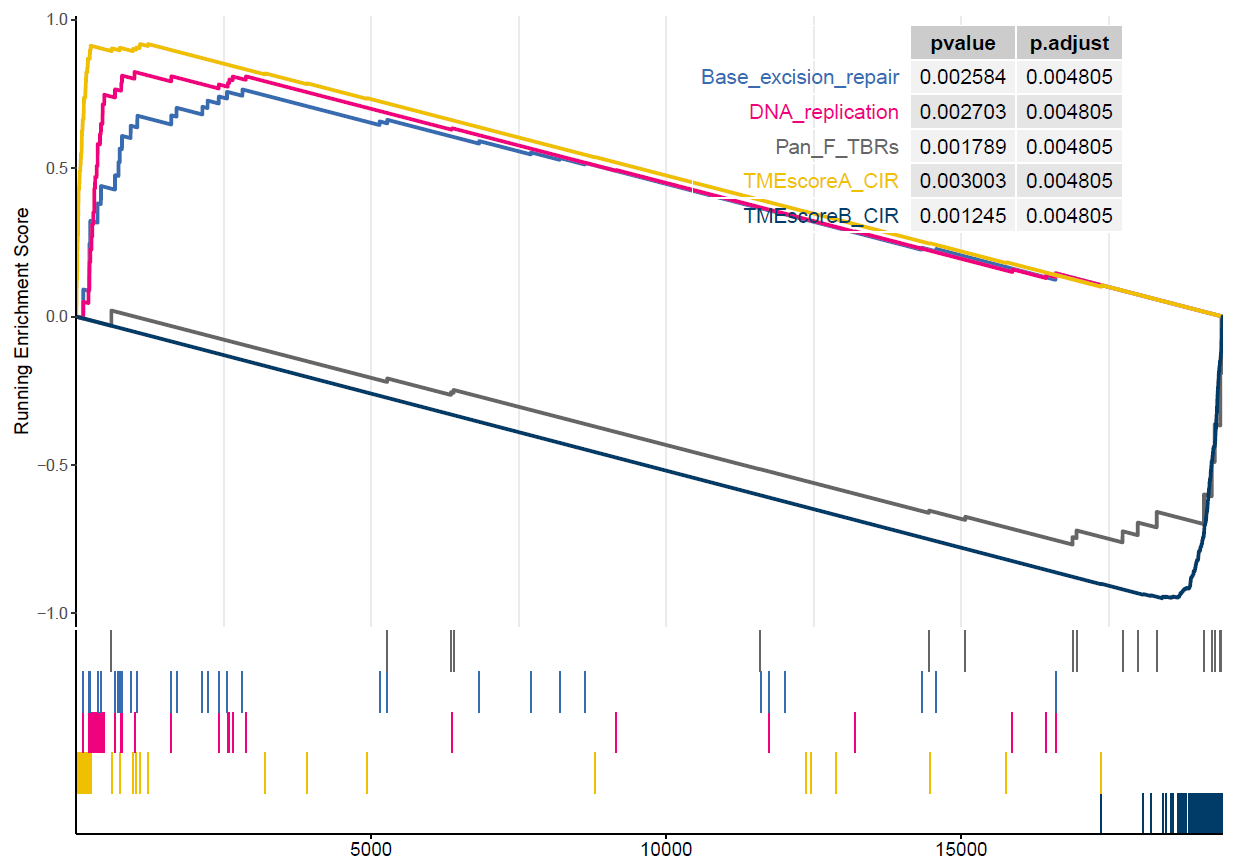
Figure 8.1: GSEA of TME gent sets
Hallmark gene signatures
gsea<- sig_gsea(deg,
genesets = NULL,
path = "GSEA",
gene_symbol = "symbol",
logfc = "log2FoldChange",
org = "hsa",
show_plot = FALSE,
msigdb = TRUE,
category = "H",
subcategory = NULL,
palette_bar = "aaas",
show_bar = 5,
show_gsea = 6)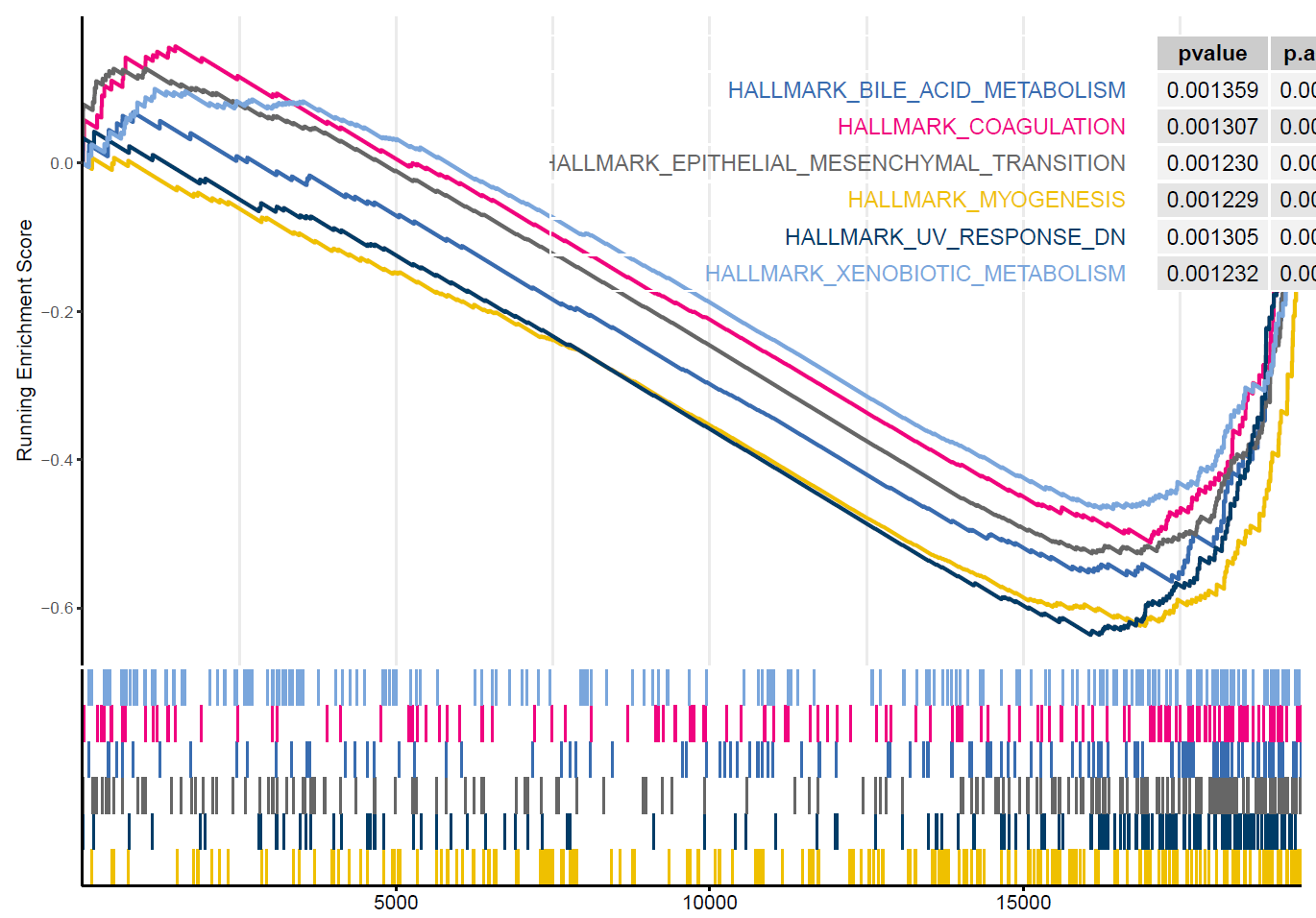
Figure 8.2: GSEA of Hallmark gent sets
8.7 DEG analysis: method2
Identifing TME subtype-related differential genes using find_markers_in_bulk
library(Seurat)
res <- find_markers_in_bulk(pdata = tme,
eset = eset,
group = "TMEcluster",
nfeatures = 2000,
top_n = 50,
thresh.use = 0.15,
only.pos = TRUE,
min.pct = 0.10)
top15 <- res$top_markers %>% dplyr:: group_by(cluster) %>% dplyr::top_n(15, avg_log2FC)
top15$gene## [1] "IDO1" "CXCL11" "SLCO1B3" "IFNG"
## [5] "AIM2" "GZMB" "VSNL1" "CXCL10"
## [9] "CXCL9" "GBP4" "KLRC2" "GNLY"
## [13] "GZMH" "KISS1R" "WARS" "C2orf40"
## [17] "OGN" "PGA4" "SCN7A" "C7"
## [21] "ADH1B" "GIF" "SCRG1" "LIPF"
## [25] "GHRL" "MAMDC2" "VIP" "ABCA8"
## [29] "ATP4A" "TMEM100" "REG1B" "MAGEA12"
## [33] "MAGEA4" "IL1A" "PI15" "IL11"
## [37] "MAGEA6" "MAGEA10-MAGEA5" "PPBP" "PROK2"
## [41] "MAGEA2B" "CLEC5A" "IL24" "CTAG1A"
## [45] "EREG"Heatmap visualisation using Seurat’s DoHeatmap
#定义分型对应的颜色
cols <- c('#2692a4','#fc0d3a','#ffbe0b')
p1 <- DoHeatmap(res$sce, top15$gene, group.colors = cols )+
scale_fill_gradientn(colours = rev(colorRampPalette(RColorBrewer::brewer.pal(11,"RdBu"))(256)))Extracting variables from the expression matrix to merge with TME subtypes
input <- combine_pd_eset(eset = eset, pdata = tme, feas = top15$gene, scale = T)
p2 <- sig_box(input, variable = "TMEcluster", signature = "IFNG", jitter = TRUE,
cols = cols, show_pvalue = TRUE, size_of_pvalue = 4)## # A tibble: 3 × 8
## .y. group1 group2 p p.adj p.format p.signif method
## <chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr>
## 1 signature IA IE 4.09e-17 1.20e-16 < 2e-16 **** Wilcoxon
## 2 signature IA IS 1.44e-13 2.90e-13 1.4e-13 **** Wilcoxon
## 3 signature IE IS 8.35e- 2 8.4 e- 2 0.084 ns Wilcoxonp3 <- sig_box(input, variable = "TMEcluster", signature = "IL1A",
jitter = TRUE, cols = cols, show_pvalue = TRUE, size_of_pvalue = 4)## # A tibble: 3 × 8
## .y. group1 group2 p p.adj p.format p.signif method
## <chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr>
## 1 signature IA IE 1.46e-10 2.90e-10 1.5e-10 **** Wilcoxon
## 2 signature IA IS 8.22e- 7 8.2 e- 7 8.2e-07 **** Wilcoxon
## 3 signature IE IS 4.90e-20 1.5 e-19 < 2e-16 **** Wilcoxonif (!requireNamespace("patchwork", quietly = TRUE)) install.packages("patchwork")
library(patchwork)
p <- (p1|p2/p3) + plot_layout(widths = c(2.3,1))
p + plot_annotation(tag_levels = 'A')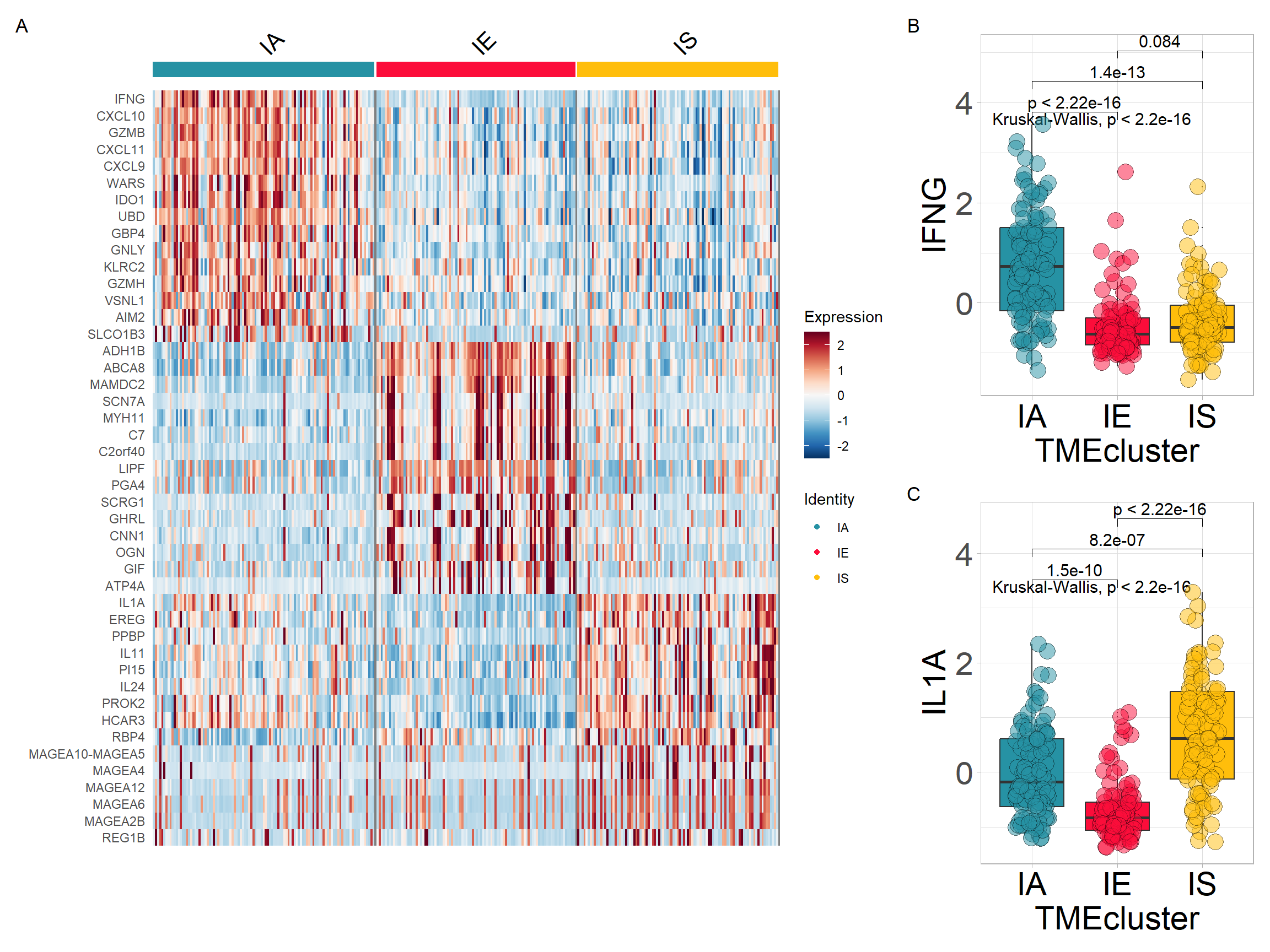
8.8 Identifying signatures associated with TME clusters
Calculate TME associated signatures-(through PCA method).
sig_tme<-calculate_sig_score(pdata = NULL,
eset = eset,
signature = signature_collection,
method = "pca",
mini_gene_count = 2)
sig_tme <- t(column_to_rownames(sig_tme, var = "ID"))
sig_tme[1:5, 1:3]## GSM1523727 GSM1523728 GSM1523729
## CD_8_T_effector -2.5513794 0.7789141 -2.1770675
## DDR -0.8747614 0.7425162 -1.3272054
## APM 1.1098368 2.1988688 -0.9516419
## Immune_Checkpoint -2.3701787 0.9455120 -1.4844104
## CellCycle_Reg 0.1063358 0.7583302 -0.3649795Finding signatures or cell types associated with TMEcluster
res <- find_markers_in_bulk(pdata = tme, eset = sig_tme, group = "TMEcluster", nfeatures = 1000, top_n = 20, min.pct = 0.10)
top15 <- res$top_markers %>% dplyr:: group_by(cluster) %>% dplyr::top_n(15, avg_log2FC)
p1 <- DoHeatmap(res$sce, top15$gene, group.colors = cols)+
scale_fill_gradientn(colours = rev(colorRampPalette(RColorBrewer::brewer.pal(11,"RdBu"))(256)))top15$gene <- gsub(top15$gene, pattern = "-", replacement = "\\_")
input <- combine_pd_eset(eset = sig_tme, pdata = tme, feas = top15$gene, scale = T)
p2 <- sig_box(input, variable = "TMEcluster", signature = "CD_8_T_effector", jitter = TRUE,
cols = cols, show_pvalue = TRUE, size_of_pvalue = 4, size_of_font = 6)## # A tibble: 3 × 8
## .y. group1 group2 p p.adj p.format p.signif method
## <chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr>
## 1 signature IA IE 3.53e-10 7.10e-10 3.5e-10 **** Wilcoxon
## 2 signature IA IS 8.49e-13 2.5 e-12 8.5e-13 **** Wilcoxon
## 3 signature IE IS 1.41e- 1 1.4 e- 1 0.14 ns Wilcoxonp3 <- sig_box(input, variable = "TMEcluster", signature = "Neutrophils_Bindea_et_al",
jitter = TRUE, cols = cols, show_pvalue = TRUE, size_of_pvalue = 4, size_of_font = 6)## # A tibble: 3 × 8
## .y. group1 group2 p p.adj p.format p.signif method
## <chr> <chr> <chr> <dbl> <dbl> <chr> <chr> <chr>
## 1 signature IA IE 0.00639 0.013 0.0064 ** Wilcoxon
## 2 signature IA IS 0.0584 0.058 0.0584 ns Wilcoxon
## 3 signature IE IS 0.0000929 0.00028 9.3e-05 **** Wilcoxon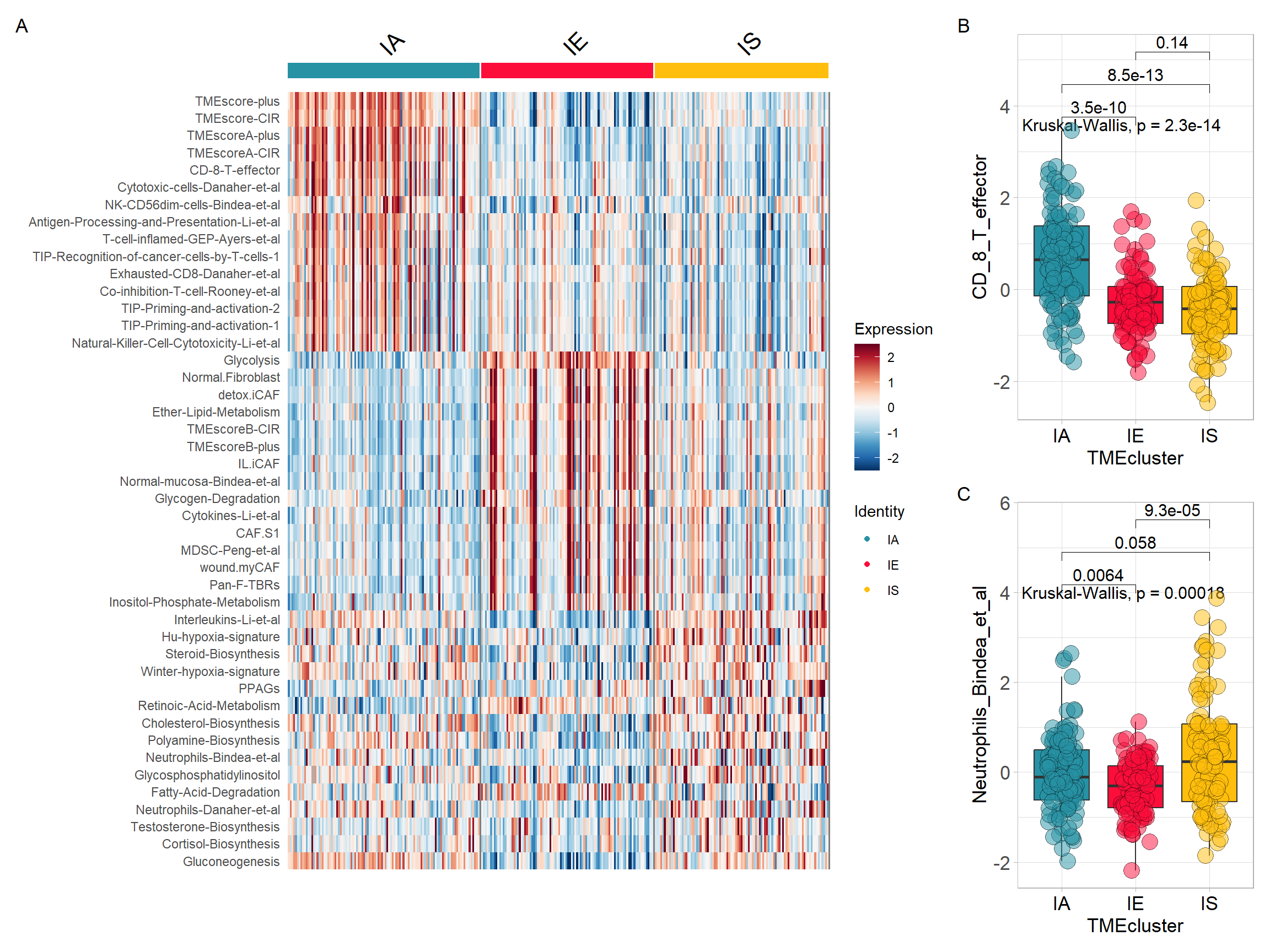
library(survminer)
data(pdata_acrg, package = "IOBR")
input <- merge(pdata_acrg, input, by = "ID")
p1<-surv_group(input_pdata = input,
target_group = "TMEcluster",
ID = "ID",
reference_group = "High",
project = "ACRG",
cols = cols,
time = "OS_time",
status = "OS_status",
time_type = "month",
save_path = "result")## >>> Dataset's survival follow up time is range between 1 to 105.7 months## IA IE IS
## 107 96 97## 1079697## Maximum of follow up time is 105.7 months; and will be divided into 6 sections;## Warning in geom_segment(aes(x = 0, y = max(y2), xend = max(x1), yend = max(y2)), : All aesthetics have length 1, but the data has 2 rows.
## ℹ Please consider using `annotate()` or provide this layer with data containing
## a single row.## Registered S3 methods overwritten by 'ggpp':
## method from
## heightDetails.titleGrob ggplot2
## widthDetails.titleGrob ggplot2## Warning in geom_segment(aes(x = 0, y = max(y2), xend = max(x1), yend = max(y2)), : All aesthetics have length 1, but the data has 2 rows.
## ℹ Please consider using `annotate()` or provide this layer with data containing
## a single row.
## All aesthetics have length 1, but the data has 2 rows.
## ℹ Please consider using `annotate()` or provide this layer with data containing
## a single row.
## All aesthetics have length 1, but the data has 2 rows.
## ℹ Please consider using `annotate()` or provide this layer with data containing
## a single row.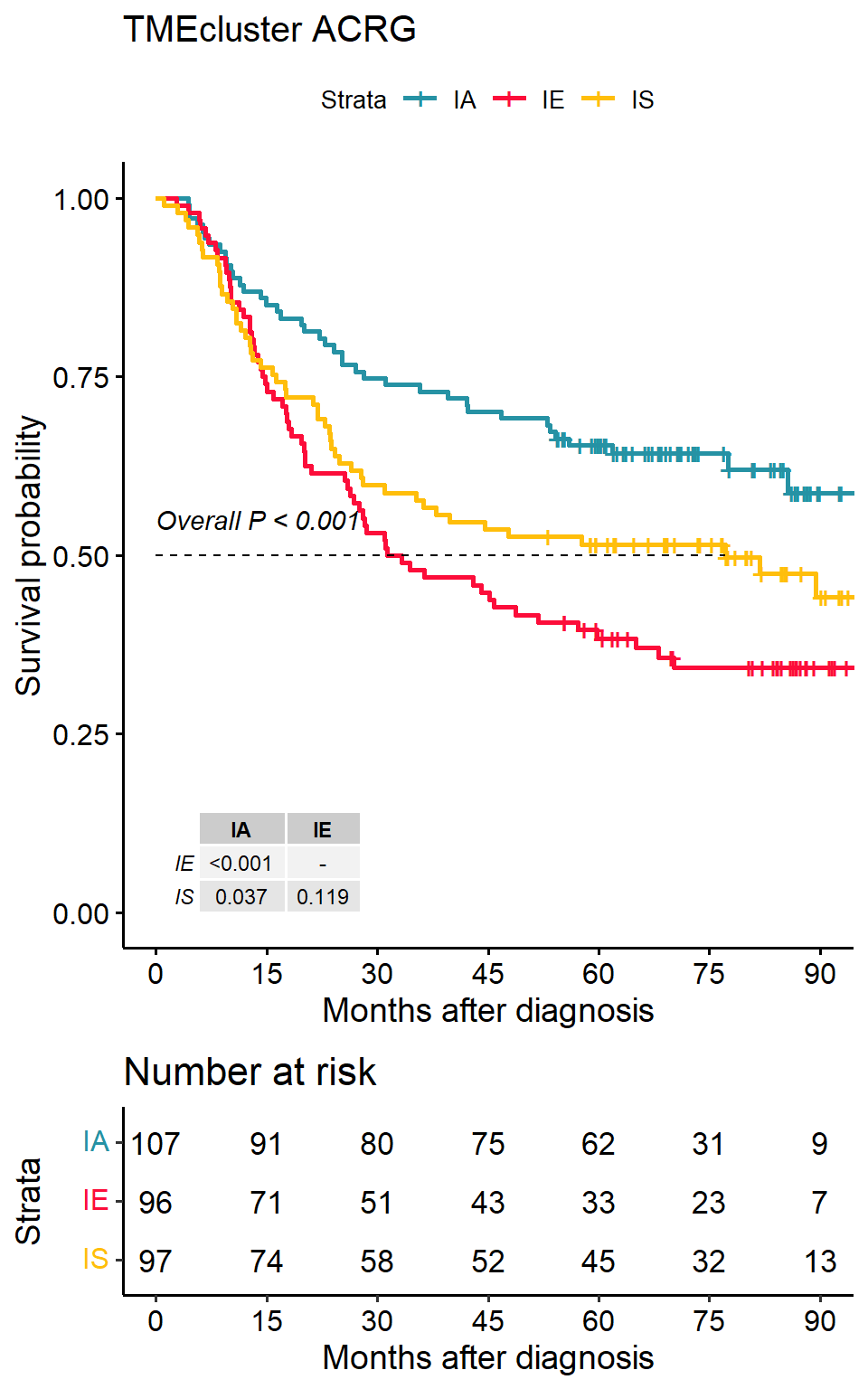
## # A tibble: 12 × 5
## # Groups: TMEcluster [3]
## TMEcluster Subtype Freq Prop count
## <chr> <fct> <dbl> <dbl> <dbl>
## 1 IA EMT 7 0.07 107
## 2 IA MSI 49 0.46 107
## 3 IA MSS/TP53- 27 0.25 107
## 4 IA MSS/TP53+ 24 0.22 107
## 5 IE EMT 24 0.25 96
## 6 IE MSI 3 0.03 96
## 7 IE MSS/TP53- 40 0.42 96
## 8 IE MSS/TP53+ 29 0.3 96
## 9 IS EMT 15 0.15 97
## 10 IS MSI 16 0.16 97
## 11 IS MSS/TP53- 40 0.41 97
## 12 IS MSS/TP53+ 26 0.27 97## [1] "'#374E55FF', '#DF8F44FF', '#00A1D5FF', '#B24745FF', '#79AF97FF', '#6A6599FF', '#80796BFF'"## # A tibble: 9 × 5
## # Groups: TMEcluster [3]
## TMEcluster Lauren Freq Prop count
## <chr> <fct> <dbl> <dbl> <dbl>
## 1 IA Diffuse 34 0.32 107
## 2 IA Intestinal 60 0.56 107
## 3 IA Mixed 13 0.12 107
## 4 IE Diffuse 60 0.62 96
## 5 IE Intestinal 32 0.33 96
## 6 IE Mixed 4 0.04 96
## 7 IS Diffuse 41 0.42 97
## 8 IS Intestinal 54 0.56 97
## 9 IS Mixed 2 0.02 97## [1] "'#374E55FF', '#DF8F44FF', '#00A1D5FF', '#B24745FF', '#79AF97FF', '#6A6599FF', '#80796BFF'"## # A tibble: 7 × 5
## # Groups: TMEcluster [3]
## TMEcluster TMEscore_binary Freq Prop count
## <chr> <fct> <dbl> <dbl> <dbl>
## 1 IA High 60 0.56 107
## 2 IA Low 47 0.44 107
## 3 IE High 5 0.05 96
## 4 IE Low 91 0.95 96
## 5 IS High 6 0.06 97
## 6 IS Low 90 0.93 97
## 7 IS <NA> 1 0.01 97## [1] "'#374E55FF', '#DF8F44FF', '#00A1D5FF', '#B24745FF', '#79AF97FF', '#6A6599FF', '#80796BFF'"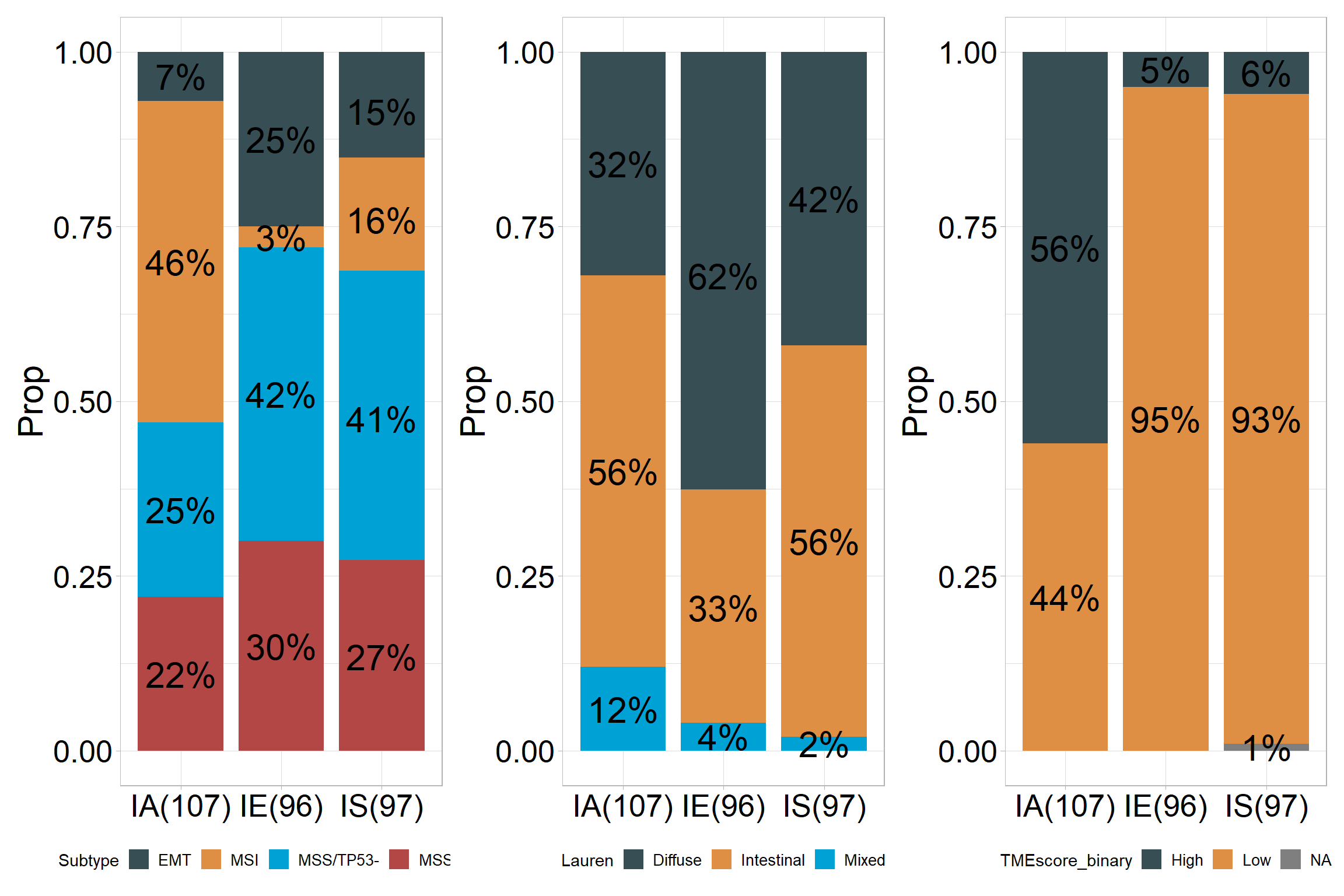
8.9 References
Cristescu, R., Lee, J., Nebozhyn, M. et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 21, 449–456 (2015). https://doi.org/10.1038/nm.3850
CIBERSORT; Newman, A. M., Liu, C. L., Green, M. R., Gentles, A. J., Feng, W., Xu, Y., … Alizadeh, A. A. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nature Methods, 12(5), 453–457. https://doi.org/10.1038/nmeth.3337;
Seurat: Hao and Hao et al. Integrated analysis of multimodal single-cell data. Cell (2021)
Zeng D, Yu Y, Qiu W, Mao Q, …, Zhang K, Liao W; Tumor microenvironment immunotyping heterogeneity reveals distinct molecular mechanisms to clinical immunotherapy applications in gastric cancer. (2024) Under Review.